Boltz-2 released on deepmirror
We have just released Boltz-2 on deepmirror. Boltz-2 is the latest in a series of molecular structure prediction models. Like AlphaFold3, Chai-1, and Boltz-1, it predicts 3D structures, but Boltz-2 also estimates binding affinity between proteins and ligands.
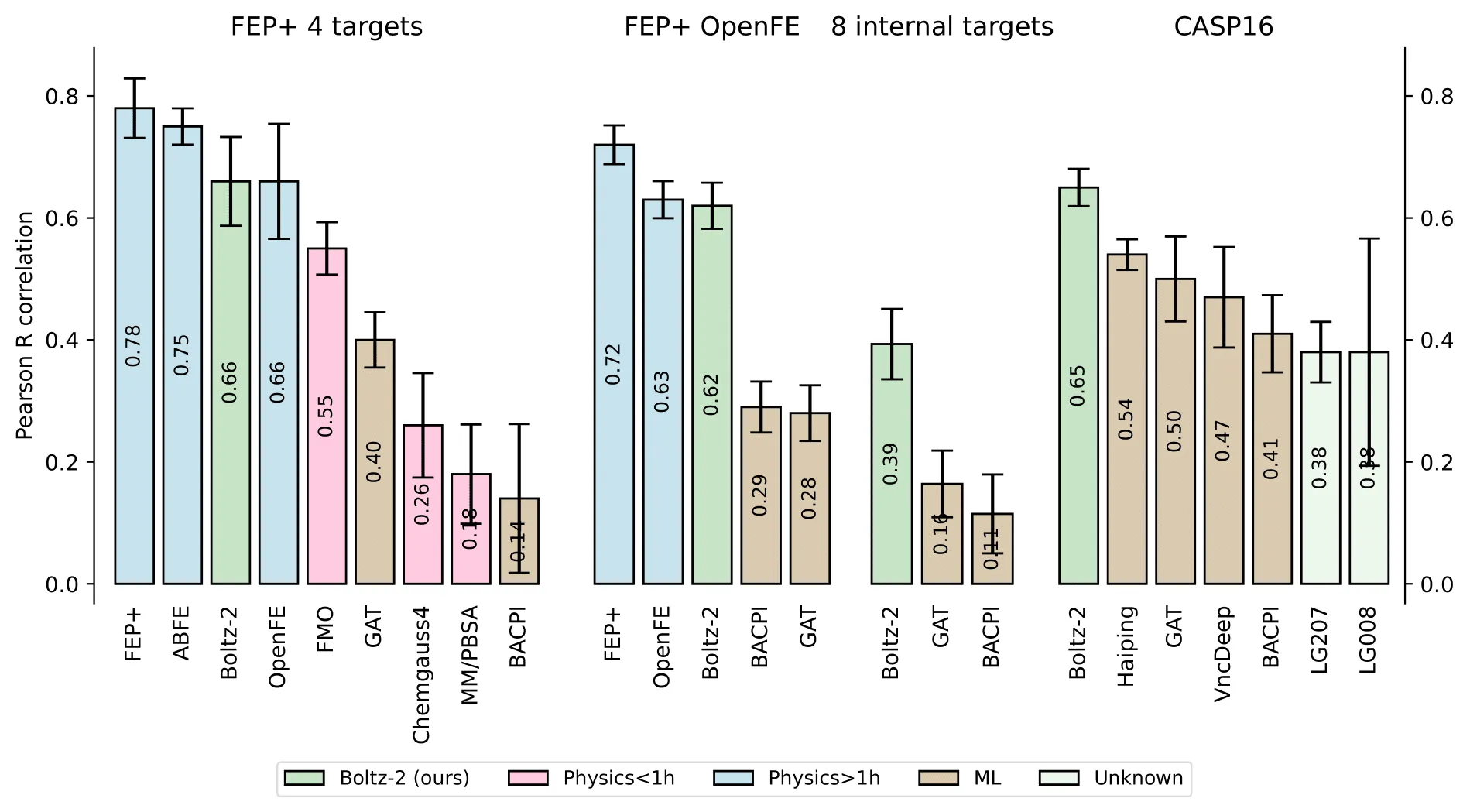
This is the first public model to combine co-folding with binding affinity predictions, while achieving speeds approximately 1000× faster than traditional physics-based energy perturbation calculations.
The team behind Boltz-2, a collaboration between MIT and Recursion, benchmarked Boltz-2 on both hit-to-lead and lead-optimization datasets. It performs well on OpenFE, with more mixed results on Recursion’s internal targets. While community evaluation is just beginning, this represents a significant step forward in scalable co-folding + affinity prediction.
You can try Boltz-2 today using the deepmirror-client and we’ve published a notebook with an example. It will also be available on the deepmirror app in the next few weeks.
If you’d like help running Boltz-2 on deepmirror, or if you need access to the platform, feel free to book a call.
References
You can find the full Boltz-2 paper at http://jeremywohlwend.com/assets/boltz2.pdf
You can find the code at https://github.com/jwohlwend/boltz
You can install the deepmirror client from https://github.com/deepmirror/deepmirror-client
.svg)


.svg)



.avif)
